After a decade of deep learning, AI is ready to make an impact on drug development. Large-scale models like AlphaFold, Evo, and ESM3 can now predict the structure of biomolecules with near experimental accuracy and design selective therapies against them. While this addresses one hurdle in drug development, most drugs still fail not because of their structure, but because they target the wrong proteins or are tested in the wrong patients. This is a fundamentally different problem and to date there are no models that tell us which targets to drug or in which patients to drug them.
This is because complex diseases, like cancer, rarely boil down to a single gene or protein. Instead, different genetic, cellular, and tissue-level processes drive disease across different patient populations, making it difficult to predict how a drug will affect a given person. For some classes of drugs, like immunotherapies which work by activating immune response, the pathways are poorly understood and the targets are mostly unknown.
To make a real impact on drug discovery, we need AI models that understand disease biology at the patient level.
Recent work has shown that large generative models, trained on multimodal data like video and text, can learn to represent aspects of the physical world and simulate complex phenomena, including those not easily observable by humans. These world models are therefore of increasing interest in domains like warehouse robotics and autonomous driving, where simulating outcomes of possible actions (in the form of realistic images and videos) can assist with decision-making and planning.
World models can also be used to simulate patient biology to inform drug discovery. To know what to target, we cannot run tests in patients – instead, we need to predict how a patient would respond to a hypothetical treatment in the context of a complex disease. Similar to other domains, simulation in biology can be used to replace trial and error by predicting the outcomes of therapeutic interactions. Here we introduce the first world model for cancer biology: the Oncology Counterfactual Therapeutics Oracle (OCTO), a discovery engine to power the next generation of cancer therapeutics.
OCTO is a massively multimodal, transformer-based model trained on data from thousands of patients' tumors generated in our wet lab specifically for self-supervised learning. The training set includes multiplex protein staining, spatial gene expression, DNA sequencing, and structural markers like H&E from each sample. These data capture complex molecular drivers and tumor-immune interactions at multiple-scales and across hundreds of millions of cells. We designed OCTO to be both highly scalable and uniquely interpretable so that it is able to leverage data and compute to improve its world model and be useful as a simulator for drug development.
OCTO is a transformer in the same vein as GPT-4o, Sora, and other large-scale generative models. It is massively multimodal, able to ingest all types of patient biological data into a single unified representation or embedding space. OCTO was trained on a joint dataset containing multiplex protein staining, spatial gene expression, DNA sequencing, and H&E. It outputs a 16-channel image capturing proteins in tumor sections in response to a visual prompt. We chose this high-dimensional, high-resolution data type as the main output for two reasons: first, because it captures therapeutically relevant features of tumor and immune biology and because it scales easily and across a wide variety of cancer biology.
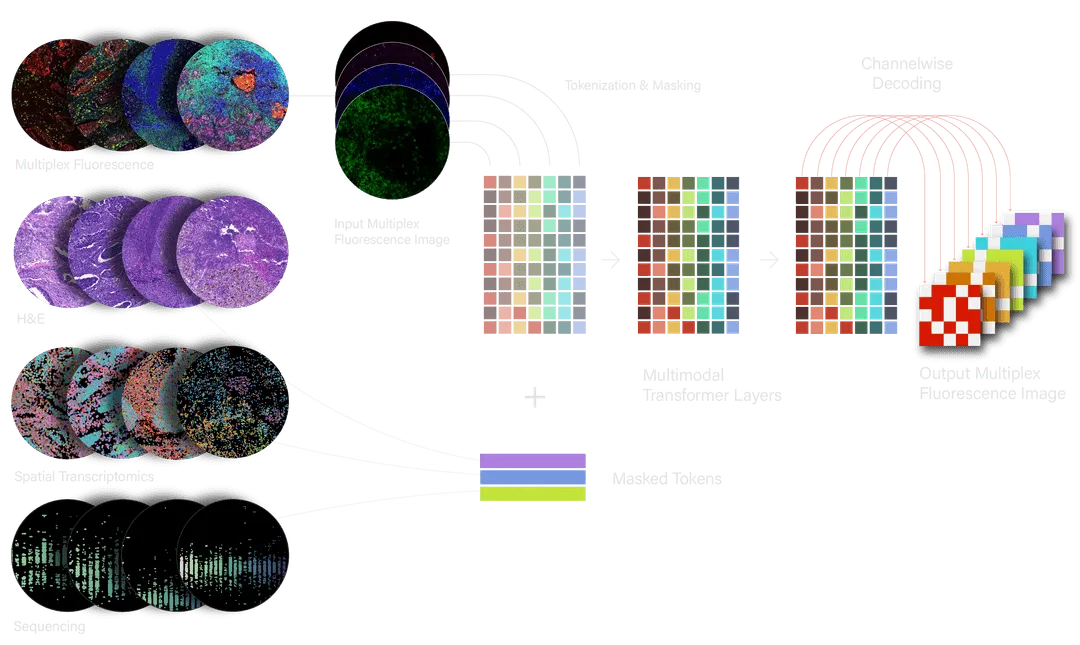
OCTO leverages the scalability of standard transformers but introduces several innovations. These are needed to make OCTO a simulator, which we can prompt with counterfactuals – “What if?” questions about how different interventions would affect patient biology.
OCTO incorporates spatially-aligned, multimodal data from different tumor regions into a unified representation. To achieve this, we encode different data types in ways that best preserve their biologically relevant features. For example, multiplex fluorescence images, which measure protein expression in cells, are first split into color channels that represent discrete proteins and then “patchified” into individual tokens. Spatial gene expression data is modeled with one token per gene at each location that contains a cell. Once processed, the tokens are combined into a single sequence so that the model can reason across modalities and learn latent relationships between the underlying biology they represent. OCTO was pre-trained on 20B tokens across 128 GPUs and is able to include up to 16,000 tokens in its “context window” at inference time.
We developed a new form of masked data modeling to train OCTO: structured multimodal masking. Standard masked image modeling masks ~75% of patches in a multichannel image, which leaves enough visible structure to infer what is missing. In contrast, OCTO is trained with nearly all tokens masked out in each of the 16 protein images and in the other data modalities. Because so little data is revealed, OCTO must learn to combine information across all modalities in a unified, spatial latent representation. This representation underlies OCTO’s ability to simulate strong relationships among sparse biological elements. For example, OCTO learns how expression levels of individual genes predict the prevalence of different cell types within a region of tumor.
To extract insights from OCTO’s internal representations and use it as a simulator, we need an easy way to query the model. We do this through a form of visual prompting in which the model is fed a partially revealed image, then infers the rest of the image. We can further predict the impact of a biological perturbation (e.g. knocking out a gene) by measuring how this change to the input data alters OCTO’s prediction. Because OCTO is trained with heavy masking, these visual prompts and perturbations may be very sparse – for instance, to simulate the altered expression of a single gene or protein.
The rest of this report expands on different ways we can prompt OCTO to extract biological knowledge. We first use simulations to confirm that OCTO has learned aspects of biology that it should learn (“ground truth”) and then show how counterfactual prompts enable in silico screening for drug targets.
After training, OCTO can accurately reconstruct high resolution, 16-channel protein images from surprisingly little data (1-2% of the total patches revealed across all protein channels).The reconstructed images are not only visually accurate but include features - such as the spatial relationships between proteins - that are biologically important but hard to detect by eye. This suggests the model has learned both a rich representation of the image as well as specific relationships between features across channels.
We can test OCTO’s knowledge of more specific biology by masking inputs in non-random ways. For instance, we see that OCTO has learned which proteins are frequently co-expressed by the same cells. When cells with one protein are revealed but all other proteins channels are masked out, OCTO renders the correct co-expressed protein (but not others) on the same cells (CD8+ T cells co-express CD3; CD163+ macrophages co-express CD68; and PanCK+ tumor cells co-express E-Cadherin.) These examples confirm that OCTO can accurately simulate biological relationships.
One type of simulation surprised us. When OCTO is prompted with images of nuclear staining alone, it accurately infers which parts of the tissue are tumor and which are immune cells. OCTO is not trained on this task explicitly; its broad self-supervised training generalizes to the more specific prompt. This is reminiscent of “emergent” zero-shot abilities in large language models. We predict that continuing to scale OCTO and its aligned, multimodal training data will yield strong performance on a wide range of therapeutically relevant tasks, even without task-specific fine tuning.
The structured masking strategy used to train OCTO is critical. This is because models learn different representations of the data depending on what is masked out and what is revealed during training. Models trained via standard masked image modeling, in which the same ~75% of patches are masked across all channels, do not learn high-level biological relationships; instead, they make predictions based on low-level image statistics. The power of specialized masking strategies is consistent with related work in the video domain, where the right masking allows models to learn about the physical structure of scenes. The space of possible masking strategies grows immense with spatially-aligned, multimodal data. Creating the strongest biological simulator will depend on finding the best strategies, including through new forms of meta-learning.
Making predictions about counterfactual data is how we assess what OCTO has learned about more subtle biological relationships. The rest of our report illustrates this with a case study of known biology, then explains how we probe unknown biology – and screen for potential drug targets – by simulating possible therapeutic scenarios.
We run counterfactual simulations by taking real data, manipulating one or more of the modalities, and feeding the perturbed inputs to OCTO. For example, we can simulate the effect of changing expression levels of a single gene on an output protein image. The difference between predictions on the original and altered inputs indicates how OCTO “thinks” expression of this gene affects levels of this protein.
Interferon gamma (IFNg) is a protein secreted by activated T cells. It has many functions in the adaptive immune response, including increasing antigen presentation and expression of the protein HLA in other cells. In the setting of cancer immunology, increasing HLA on tumor cells makes them more susceptible to attack by the immune system.
We asked whether OCTO knows about this relationship between IFNg and HLA. Gene expression is just another of OCTO's masked inputs, so simulations behave similarly to those with protein expression. Since the expression of a number of genes is stimulated by IFNg, we simulate changes in IFNg signaling by counterfactually increasing or decreasing expression of this gene panel. Given these altered gene expression patterns, OCTO does in fact predict the expected, dose-dependent changes in tumor HLA protein.
There are also hints that OCTO has learned more subtle biology. Small simulated increases in IFNg signaling raise HLA levels at the tumor border but decrease them in the interior – a spatial pattern that we began to notice in many patients after seeing these predictions. This shows how simulation with a world model can reveal structure in the data that would be hard to detect in aggregate analysis.
OCTO’s training data includes gene expression data for an increasing set of potential drug targets and therapeutically relevant pathways, all spatially aligned to other data layers. Multimodal counterfactual simulation allows us to ask: for each molecular target, what would happen to specific cohorts of patient tumors if this target were perturbed?
These in silico screens allow us to find the targets most strongly associated with a therapeutically desirable effect, such as increased engagement of immune cells with a tumor. Targets that rank highly are advanced to downstream validation: we test the impact of perturbing them in an in vivo experimental system. Besides measuring OCTO’s accuracy as a simulator, results of these validation experiments are a direct learning signal. Since OCTO can be conditioned just as well on “actions” of target perturbation as on purely observational data, this feedback can improve the model (analogous to RLHF with language models.)
Finding the right therapies for the right patients requires a new way of thinking about drug discovery. Most clinical trials fail because initially promising results do not generalize to human disease and because there are few one-size-fits-all treatments. OCTO, as a world model of patient biology, gives us an unprecedented ability to simulate patient-specific treatments and to understand cancer diversity more broadly. Today, we apply these simulations at the level of cohorts of patients, to discover precision therapies that could apply to well defined subsets of patients. However, OCTO can also operate at the level of individual patients to give us a more nuanced understanding of therapeutic efficacy.
This enables our larger vision of a rapid, positive feedback loop: (1) simulating patient biology from massively multimodal data, (2) extracting predictions to propose therapeutic actions, (3) testing these actions in a proprietary experimental in vivo system, and ultimately (4) using clinical outcomes as input data for further learning. We are enacting this loop at an unprecedented scale, not within a highly reduced model system, but directly at the level of patient biology in all of its complexity.













